Make Academic Writing Easy
Choose WriteWise for writing and editing
any type of academic text
Make Academic
Writing Easy
Choose WriteWise for writing and editing
any type of academic text
Make Academic
Writing Easy
Choose WriteWise for writing and editing
any type of academic text

Used by Thousands of Researchers & Universities











Used by Hundreds of Researchers & Universities








Used by Hundreds of Researchers & Universities





We Transform Academic Writing
Use AI to go from draft to paper in a flash
ChatGPT-4 for Rapid Writing
Get the best out of your ideas by using the best AI-writing tool: ChatGPT-4

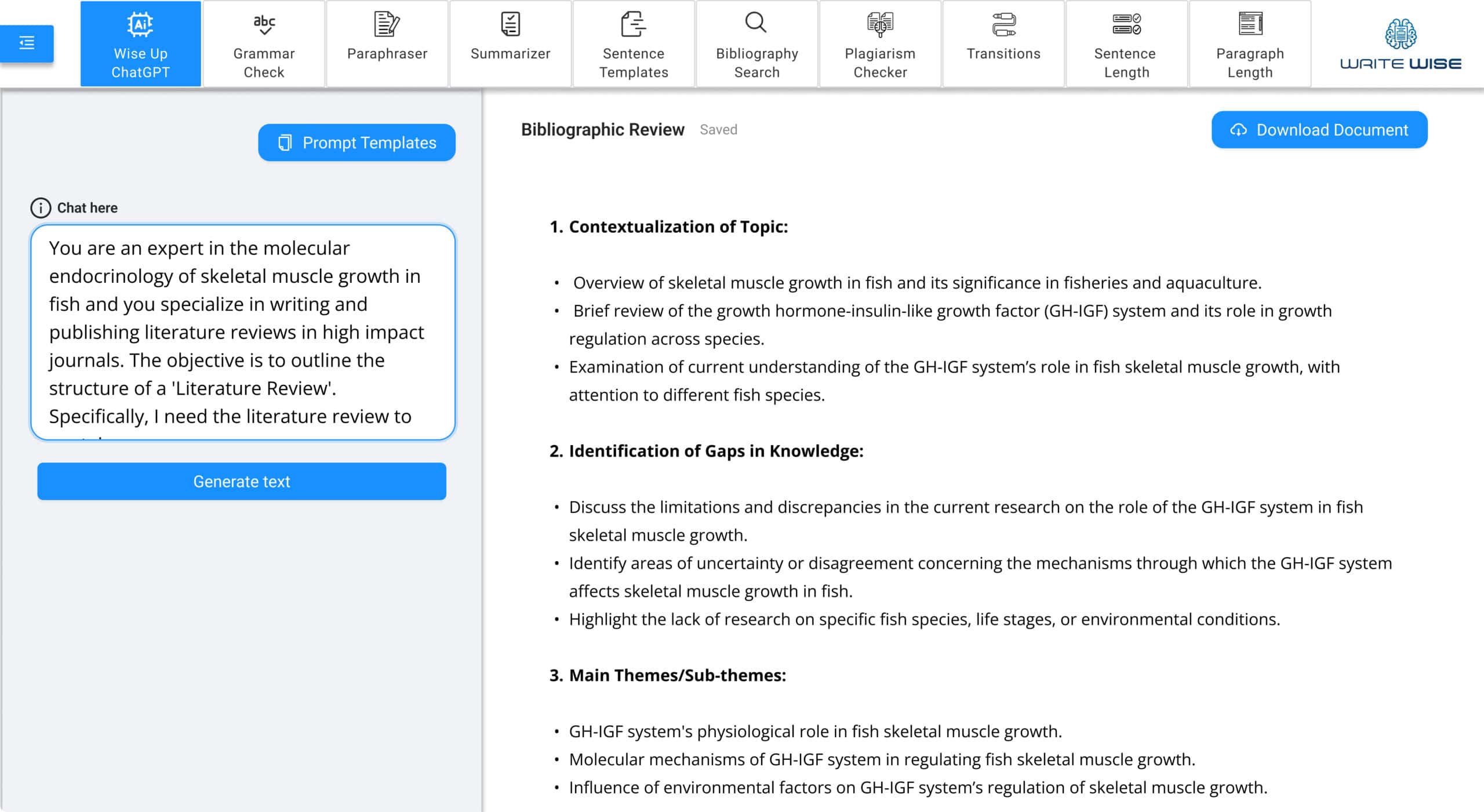
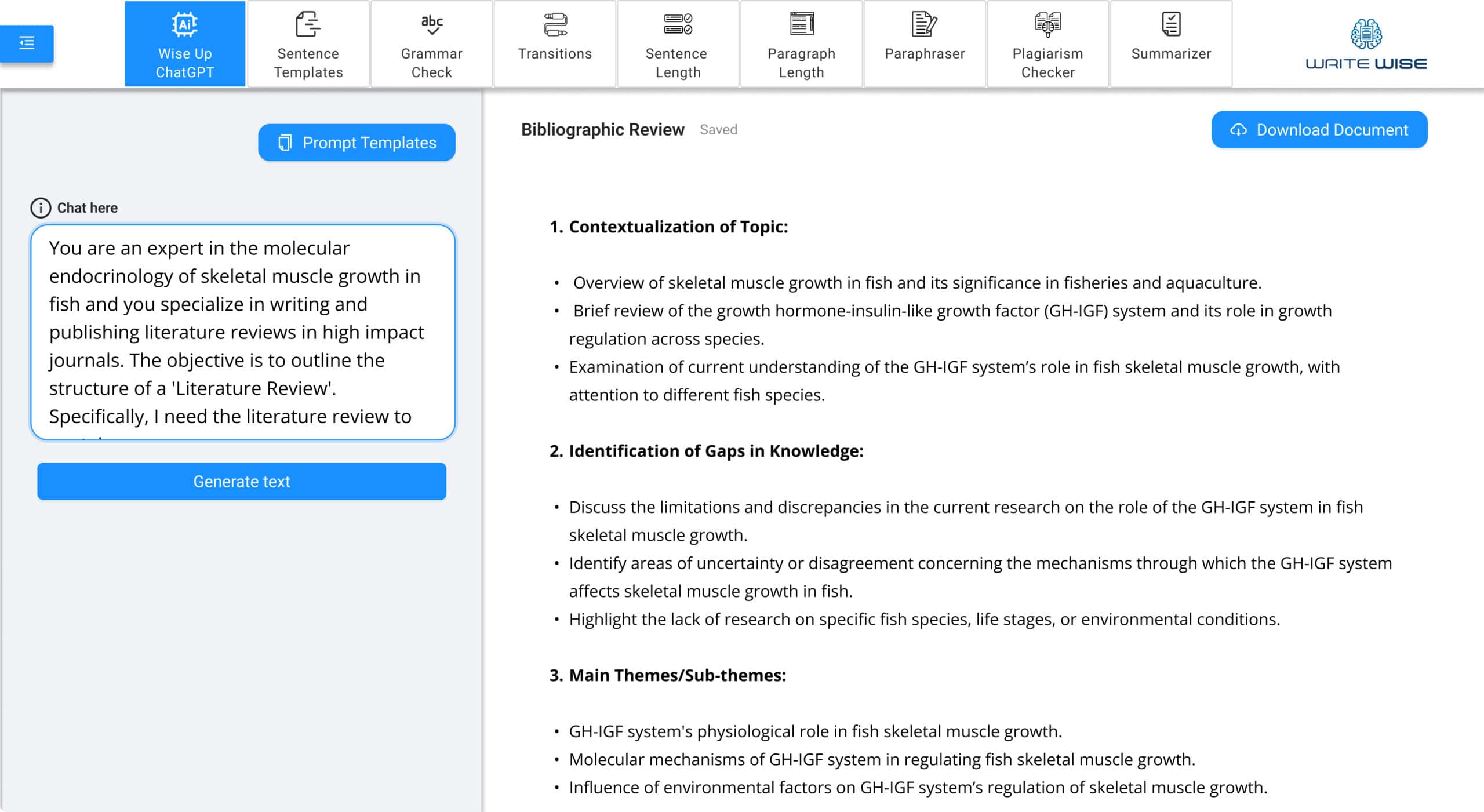
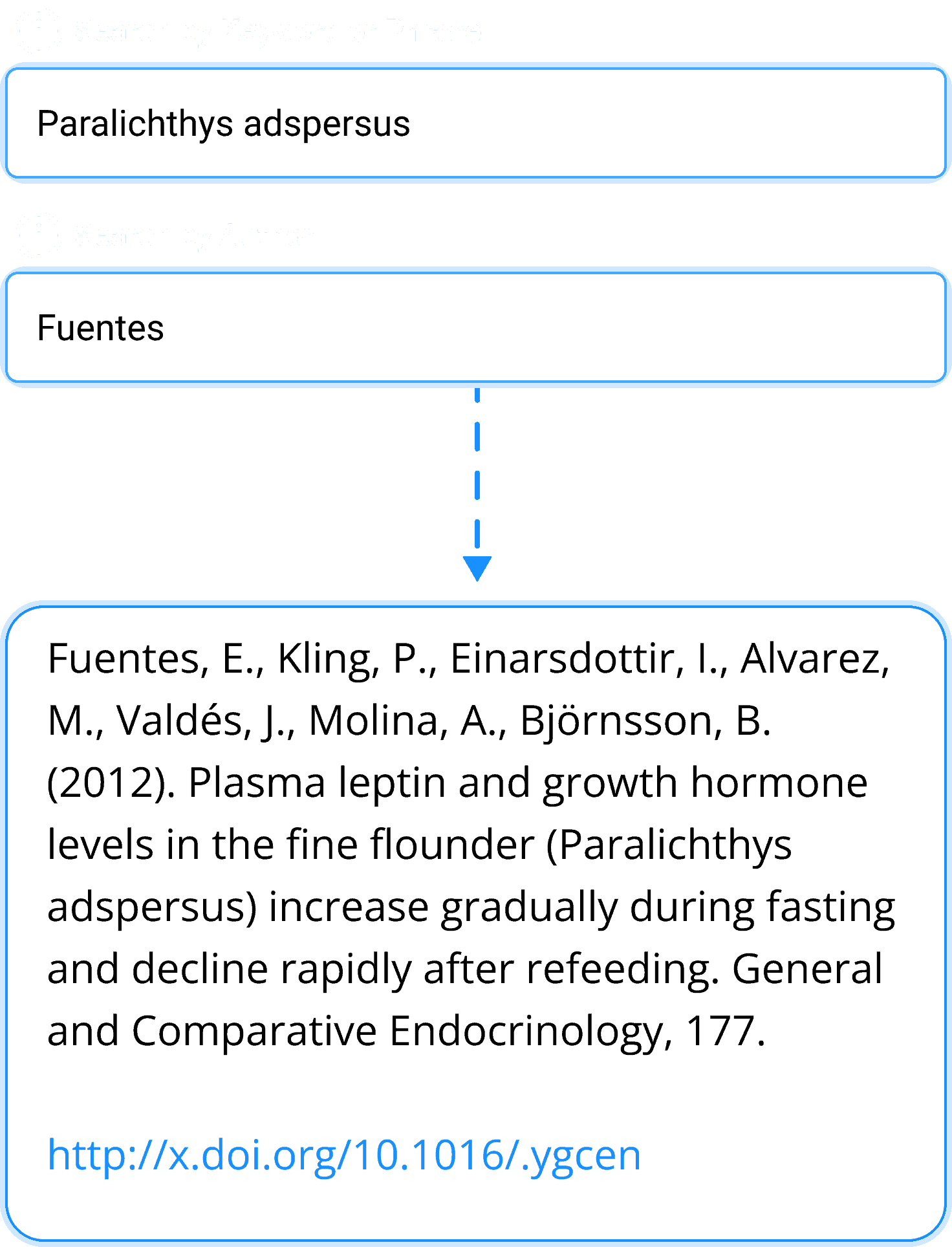
Bibliography Search
Quickly search thousands of manuscripts to reference for your project or paper
Easily and Quickly Write
High Quality Academic Texts
AI Grammar Check
Harness the power of an OpenAI GPT to intelligently check for grammatical and structural errors.
Plagiarism Detector
Verify the originality of your entire text in just one click, and get bibliographic citations when plagiarism is found.
AI Summarizer
Cut down long texts in an instant with an AI-driven summarizer. Reduce wordiness in just three clicks.
Template Prompts
Don’t know where to begin with ChatGPT-4? Use one of thousands of prompt templates to get started.
AI Paraphraser
Paraphrase any phrase, sentence, or paragraph in your academic text or project with ease.
Comprehension
Use the transitions checker and sentence/paragraph length reviewer to ensure high reader comprehension.
We Transform Academic Writing
Use AI to go from draft to paper in a flash
ChatGPT-4 for Rapid Writing
Get the best out of your ideas by using the best AI-writing tool: ChatGPT-4

Bibliography Search
Quickly search thousands of manuscripts to reference for your project or paper

Easily and Quickly Write
High Quality Academic Texts
AI Grammar Check
Harness the power of an OpenAI GPT to intelligently check for grammatical and structural errors.
Plagiarism Detector
Verify the originality of your entire text in just one click, and get bibliographic citations when plagiarism is found.
AI Summarizer
Cut down long texts in an instant with an AI-driven summarizer. Reduce wordiness in just three clicks.
Template Prompts
Don’t know where to begin with ChatGPT-4? Use one of thousands of prompt templates to get started.
AI Paraphraser
Paraphrase any phrase, sentence, or paragraph in your academic text or project with ease.
Comprehension
Use the transitions checker and sentence/paragraph length reviewer to ensure high reader comprehension.
We Transform Academic Writing
Use AI to go from draft to paper in a flash
ChatGPT-4 for Rapid Writing
Get the best out of your ideas by using the best AI-writing tool: ChatGPT-4

Bibliography Search
Quickly search thousands of manuscripts to reference for your project or paper

Easily and Quickly Write
High Quality Academic Texts
AI Grammar Check
Harness the power of an OpenAI GPT to intelligently check for grammatical and structural errors.
Plagiarism Detector
Verify the originality of your entire text in just one click, and get bibliographic citations when plagiarism is found.
AI Summarizer
Cut down long texts in an instant with an AI-driven summarizer. Reduce wordiness in just three clicks.
Template Prompts
Don’t know where to begin with ChatGPT-4? Use one of thousands of prompt templates to get started.
AI Paraphraser
Paraphrase any phrase, sentence, or paragraph in your academic text or project with ease.
Comprehension
Use the transitions checker and sentence/paragraph length reviewer to ensure high reader comprehension.
Supported by


Supported by


Supported by


Testimonials
Writing with confidence and getting results




















National & International Awards





National & International
Awards





National & International
Awards





Start Today and Boost
Your Writing Potential
WriteWise is the n°1 trusted tool for academic writing.
This is the program you’ve been searching for to write.
Start Today and Boost
Your Writing Potential
WriteWise is the n°1 trusted tool for academic writing.
This is the program you’ve been searching for to write.
Start Today and Boost
Your Writing Potential
WriteWise is the n°1 trusted tool for academic writing.
This is the program you’ve been searching for to write.


